Smart sEMG Averaging prior to synergy extraction
Post: 2020.11.13 10:45
General description
The following algorithm includes code for the segmentation of independant muscle activation from sEMG data recorded during periodic tasks. Technical details were published here. The code requires a priori segmentation of the individual muscle activation on each channel. In the following lines, a sample script will be run using test data. Details about data formatting will be provided so other authors can adapt their data to the script provided.
Data format
Current code requires data files containing sEMG signals recorded during periodic motion. Moreover, each file must contain information about data sampling rate and segmentation samples of each individual muscle activation.
Each data file must be a ‘.mat’ structure named ‘output’ with the following format:
>> output.emgData
The field ‘emgData’ contains the NxM data matrix where N is the number of channels and M is the number of samples of each channel. The data recorded should be a periodic motion repeated at a relatively stable speed.
>> output.conf.register.fm
>> output.conf.register.muscle_names
The field ‘register’ within ‘conf’ should contain a variable ‘fm’ with the sampling frequency as an integer. In addition the field output.conf.register.muscle_names contain a vector of cells with the names of each muscle. This last variable is not necessary for running the code.
>> output.processing.pattern.minims
>> output.processing.pattern.maxims
The variable maxims is NxC matrix where N is the number of channels and C the number of repetitions of the task. Each value represents the point in which the maximun peack of the averaged muscle activation occurred. The variable minims is an Nx(C+1) matrix that contains the starting and ending positions of each muscle activation. Following figure shows the maxima and minima of a single channel.
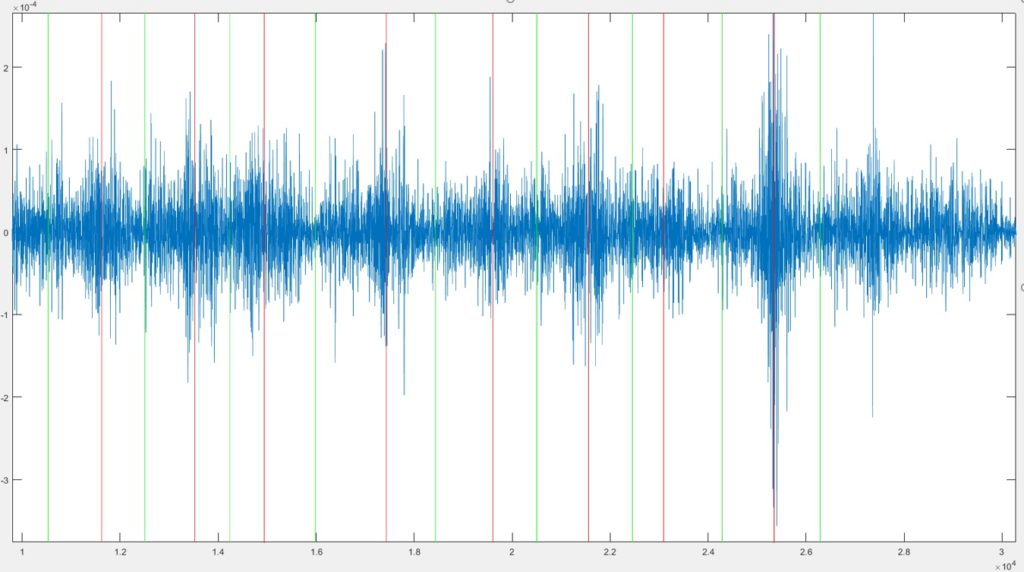
The first and last minima do not need to correspond with the first and last sample of the signal. Only data contained between these two minima will be evaluated by the code.
Sample material
Download the material required to run this code on the following link
Sample data description
The sample data set consist of 8 files containing each one 8 repetitions of elbow extension/flexion task. Each file contains EMG data from 11 electrodes located in the right upper limb with the following distribution: wrist flexor, pronator teres, wrist extensor, biceps, triceps, anterior deltoid, medium deltoid, posterior deltoid, pectoralis major, trapezius and infra spinatus.
Script description
Script Name: AveragingData.m
Script Description: This script has been tested in matlab version R2019a on a computer running windows 10. Functions required to run the script can be found in the library folder from the sample material.
function AvgData = AveragingData
% Matlab version R2019a.
% Windows 10.
% Choose the files containing the EMG data.
fprintf(‘Choose data files:’)
files = get_file_names(‘mat’,1);
% Combine all files in a single variable.
[Data] = combineEmgTrials(files);
% Obtain the average trial from the data variable.
[avgTrial,struct1] = motionModel3(Data);
% Plot Averaged Data.
figure;plot(avgTrial’)
Running the script: Step-by-Step
1) Download sample material.
2) Set the matlab workspace into the folder AveragingData.

3) Add to the path the folder “Library“.

4) Run the code ‘AvgData = AveragingScript‘.

5) The script asks for EMG files. Choose all the files from the folder ‘Data’.

6) After the script run the average trial will be plotted and storage in the variable ‘AvgData’.
References
Á. Costa-García et al., “Segmentation and Averaging of sEMG Muscle Activations Prior to Synergy Extraction,” in IEEE Robotics and Automation Letters, vol. 5, no. 2, pp. 3106-3112, April 2020, doi: 10.1109/LRA.2020.2975729.
Related Entries
Smart sEMG Averaging prior to synergy extraction
General description The following algorithm includes code for the segmentation of independant muscle activation from sEMG data recorded during periodic tasks. Technical details were published here. The code requires a priori segmentation of the individual muscle activation on each channel. In the following lines, a sample script will be run using test data. Details about […]
Corticospinal Decoding from EEG
General Description The following algorithm computes a corticospinal decoding of motor primitives from EEG signals after applying a cross-validation (one-file out) with all the recording input files. Data format Data fields description: data.EEG – contains a matrix with filtered EEG signals after artifact removal from all 16 recorded channels (27 seconds) data.EMG – contains a […]